Function to simulate a key innovation in macro-evolution with the innovative clade decoupling from the diversity-dependent diversification dynamics of the main clade
dd_KI_sim.RdSimulating a diversity-dependent diversification process where at a given time a new clade emerges with different inherent speciation rate and extinction rate and clade-level carrying capacity and with decoupled dynamics
Arguments
- pars
Vector of parameters:
pars[1]corresponds to lambda_M (speciation rate of the main clade)pars[2]corresponds to mu_M (extinction rate of the main clade)pars[3]corresponds to K_M (clade-level carrying capacity of the main clade)pars[4]corresponds to lambda_S (speciation rate of the subclade)pars[5]corresponds to mu_S (extinction rate of the subclade)pars[5]corresponds to K_S (clade-level carrying capacity of the subclade)pars[7]tinn, the time the shift in rates occurs in the lineage leading to the subclade- age
Sets the crown age for the simulation
- ddmodel
Sets the model of diversity-dependence:
ddmodel == 1: linear dependence in speciation rate with parameter K (= diversity where speciation = extinction)ddmodel == 1.3: linear dependence in speciation rate with parameter K' (= diversity where speciation = 0)ddmodel == 2: exponential dependence in speciation rate with parameter K (= diversity where speciation = extinction)ddmodel == 2.1: variant of exponential dependence in speciation rate with offset at infinityddmodel == 2.2: 1/n dependence in speciation rateddmodel == 2.3: exponential dependence in speciation rate with parameter x (= exponent)ddmodel == 3: linear dependence in extinction rateddmodel == 4: exponential dependence in extinction rateddmodel == 4.1: variant of exponential dependence in extinction rate with offset at infinityddmodel == 4.2: 1/n dependence in extinction rate with offset at infinity
Value
- out
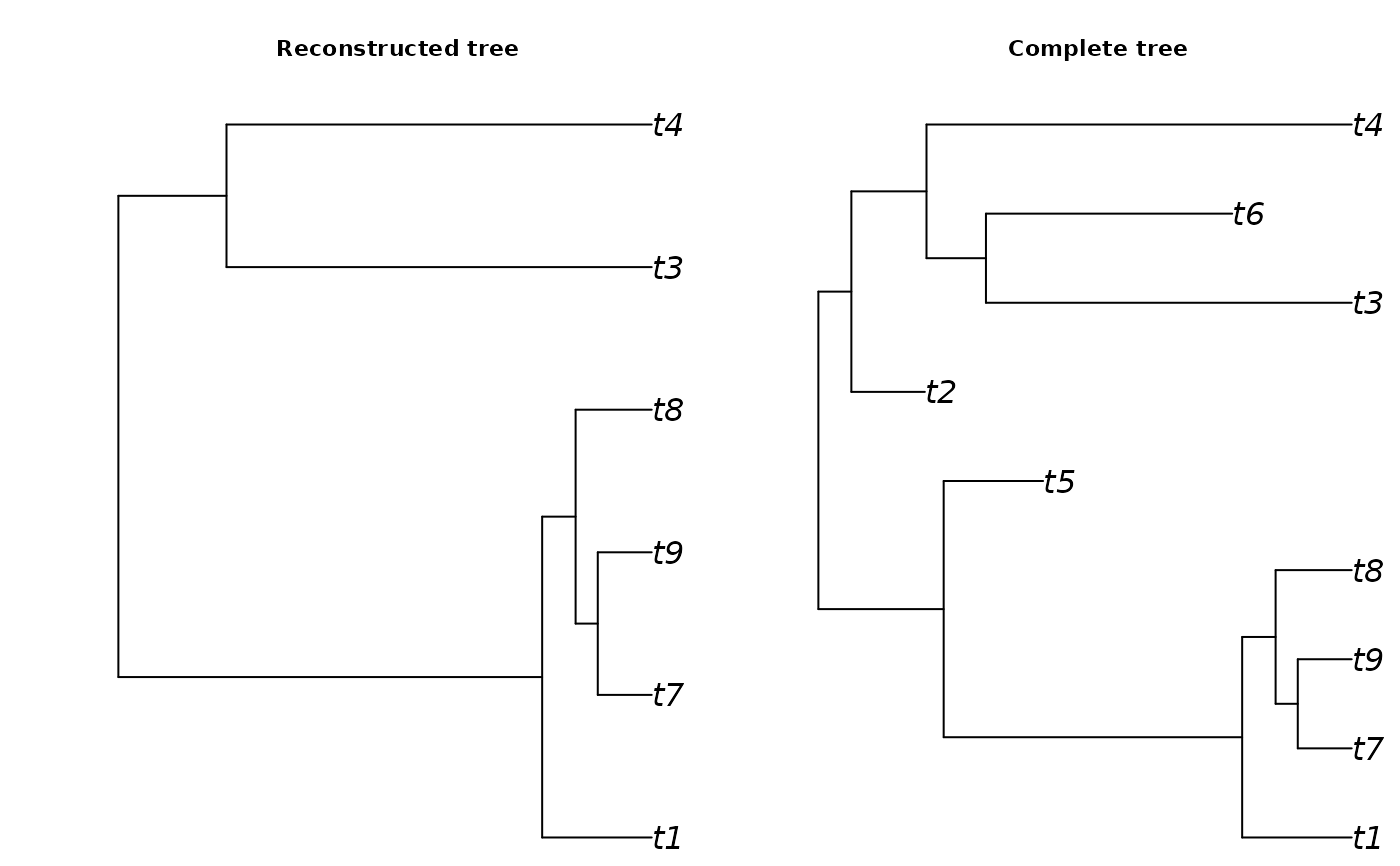
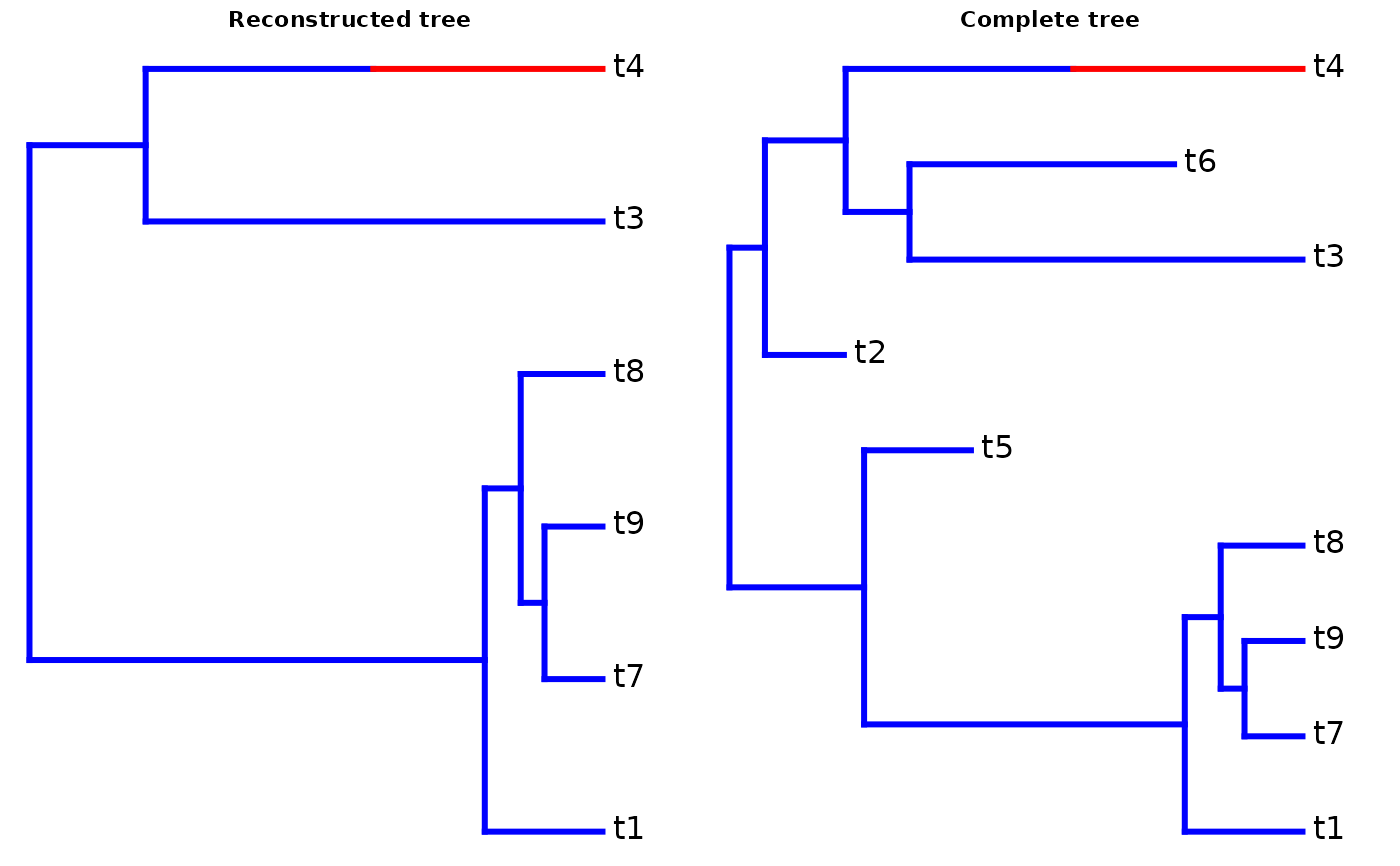
A list with the following elements: The first element is the tree of extant species in phylo format
The second element is the tree of all species, including extinct species, in phylo format
The third element is a matrix of all species where
- the first column is the time at which a species is born
- the second column is the label of the parent of the species; positive and negative values only indicate whether the species belongs to the left or right crown lineage
- the third column is the label of the daughter species itself; positive and negative values only indicate whether the species belongs to the left or right crown lineage
- the fourth column is the time of extinction of the species
If the fourth element equals -1, then the species is still extant.
- the fifth column indicates whether the species belong to the main clade (0) or the subclade (1)
The fourth element is the subclade tree of extant species (without stem)
The fifth element is the subclade tree of all species (without stem)
The sixth element is the same as the first, except that it has attributed 0 for the main clade and 1 for the subclade
The seventh element is the same as the Second, except that it has attributed 0 for the main clade and 1 for the subclade
The sixth and seventh element will be NULL if the subclade does not exist (because it went extinct).
References
- Etienne, R.S. et al. 2012, Proc. Roy. Soc. B 279: 1300-1309,
doi: 10.1098/rspb.2011.1439
- Etienne, R.S. & B. Haegeman 2012. Am. Nat.
180: E75-E89, doi: 10.1086/667574
Examples
dd_KI_sim(c(0.2,0.1,20,0.1,0.05,30,4),10)
 #> $tes
#>
#> Phylogenetic tree with 8 tips and 7 internal nodes.
#>
#> Tip labels:
#> t1, t9, t10, t8, t6, t13, ...
#>
#> Rooted; includes branch length(s).
#>
#> $tas
#>
#> Phylogenetic tree with 13 tips and 12 internal nodes.
#>
#> Tip labels:
#> t1, t9, t10, t8, t2, t4, ...
#>
#> Rooted; includes branch length(s).
#>
#> $L
#> [,1] [,2] [,3] [,4] [,5]
#> [1,] 10.00000000 0 -1 -1.0000000 0
#> [2,] 10.00000000 -1 2 6.0324077 0
#> [3,] 7.71544830 2 3 2.7422639 0
#> [4,] 7.33115475 2 4 4.3769175 0
#> [5,] 7.26533859 3 5 -1.0000000 0
#> [6,] 5.44666769 4 6 -1.0000000 1
#> [7,] 5.04996898 3 7 1.0552860 0
#> [8,] 3.88506468 -1 -8 -1.0000000 0
#> [9,] 2.84387093 -1 -9 -1.0000000 0
#> [10,] 2.39720551 -9 -10 -1.0000000 0
#> [11,] 0.84916460 5 11 -1.0000000 0
#> [12,] 0.76349279 11 12 0.1043106 0
#> [13,] 0.09764081 6 13 -1.0000000 1
#>
#> $tesS
#>
#> Phylogenetic tree with 2 tips and 1 internal node.
#>
#> Tip labels:
#> t6, t13
#>
#> Rooted; includes branch length(s).
#>
#> $tasS
#>
#> Phylogenetic tree with 2 tips and 1 internal node.
#>
#> Tip labels:
#> t6, t13
#>
#> Rooted; includes branch length(s).
#>
#> $tes2
#>
#> Phylogenetic tree with 8 tips and 7 internal nodes.
#>
#> Tip labels:
#> t1, t9, t10, t8, t6, t13, ...
#>
#> The tree includes a mapped, 2-state discrete character
#> with states:
#> 0, 1
#>
#> Rooted; includes branch lengths.
#>
#> $tas2
#>
#> Phylogenetic tree with 13 tips and 12 internal nodes.
#>
#> Tip labels:
#> t1, t9, t10, t8, t2, t4, ...
#>
#> The tree includes a mapped, 2-state discrete character
#> with states:
#> 0, 1
#>
#> Rooted; includes branch lengths.
#>
#> $tes
#>
#> Phylogenetic tree with 8 tips and 7 internal nodes.
#>
#> Tip labels:
#> t1, t9, t10, t8, t6, t13, ...
#>
#> Rooted; includes branch length(s).
#>
#> $tas
#>
#> Phylogenetic tree with 13 tips and 12 internal nodes.
#>
#> Tip labels:
#> t1, t9, t10, t8, t2, t4, ...
#>
#> Rooted; includes branch length(s).
#>
#> $L
#> [,1] [,2] [,3] [,4] [,5]
#> [1,] 10.00000000 0 -1 -1.0000000 0
#> [2,] 10.00000000 -1 2 6.0324077 0
#> [3,] 7.71544830 2 3 2.7422639 0
#> [4,] 7.33115475 2 4 4.3769175 0
#> [5,] 7.26533859 3 5 -1.0000000 0
#> [6,] 5.44666769 4 6 -1.0000000 1
#> [7,] 5.04996898 3 7 1.0552860 0
#> [8,] 3.88506468 -1 -8 -1.0000000 0
#> [9,] 2.84387093 -1 -9 -1.0000000 0
#> [10,] 2.39720551 -9 -10 -1.0000000 0
#> [11,] 0.84916460 5 11 -1.0000000 0
#> [12,] 0.76349279 11 12 0.1043106 0
#> [13,] 0.09764081 6 13 -1.0000000 1
#>
#> $tesS
#>
#> Phylogenetic tree with 2 tips and 1 internal node.
#>
#> Tip labels:
#> t6, t13
#>
#> Rooted; includes branch length(s).
#>
#> $tasS
#>
#> Phylogenetic tree with 2 tips and 1 internal node.
#>
#> Tip labels:
#> t6, t13
#>
#> Rooted; includes branch length(s).
#>
#> $tes2
#>
#> Phylogenetic tree with 8 tips and 7 internal nodes.
#>
#> Tip labels:
#> t1, t9, t10, t8, t6, t13, ...
#>
#> The tree includes a mapped, 2-state discrete character
#> with states:
#> 0, 1
#>
#> Rooted; includes branch lengths.
#>
#> $tas2
#>
#> Phylogenetic tree with 13 tips and 12 internal nodes.
#>
#> Tip labels:
#> t1, t9, t10, t8, t2, t4, ...
#>
#> The tree includes a mapped, 2-state discrete character
#> with states:
#> 0, 1
#>
#> Rooted; includes branch lengths.
#>